High-Throughput Phenotyping with EHR using a Common Automated Pipeline
PheCAP-package.RdImplement surrogate-assisted feature extraction (SAFE) and common machine learning approaches to train and validate phenotyping models. Background and details about the methods can be found at Zhang et al. (2019) <doi:10.1038/s41596-019-0227-6>, Yu et al. (2017) <doi:10.1093/jamia/ocw135>, and Liao et al. (2015) <doi:10.1136/bmj.h1885>.
Details
The DESCRIPTION file:
This package was not yet installed at build time.
Index: This package was not yet installed at build time.
PheCAP provides a straightforward interface for conducting
phenotyping on eletronic health records. One can specify the
data via PhecapData, define surrogate using
PhecapSurrogate. Next, one may run
surrogate-assisted feature extraction (SAFE) by calling
phecap_run_feature_extraction, and then
train and validate phenotyping models via
phecap_train_phenotyping_model and
phecap_validate_phenotyping_model.
The predictive performance can be visualized using
phecap_plot_roc_curves.
Predicted phenotype is provided by
phecap_predict_phenotype.
Author
NA
Maintainer: NA
References
Yu, S., Chakrabortty, A., Liao, K. P., Cai, T., Ananthakrishnan, A. N., Gainer, V. S., ... & Cai, T. (2016). Surrogate-assisted feature extraction for high-throughput phenotyping. Journal of the American Medical Informatics Association, 24(e1), e143-e149.
Liao, K. P., Cai, T., Savova, G. K., Murphy, S. N., Karlson, E. W., Ananthakrishnan, A. N., ... & Churchill, S. (2015). Development of phenotype algorithms using electronic medical records and incorporating natural language processing. bmj, 350, h1885.
Examples
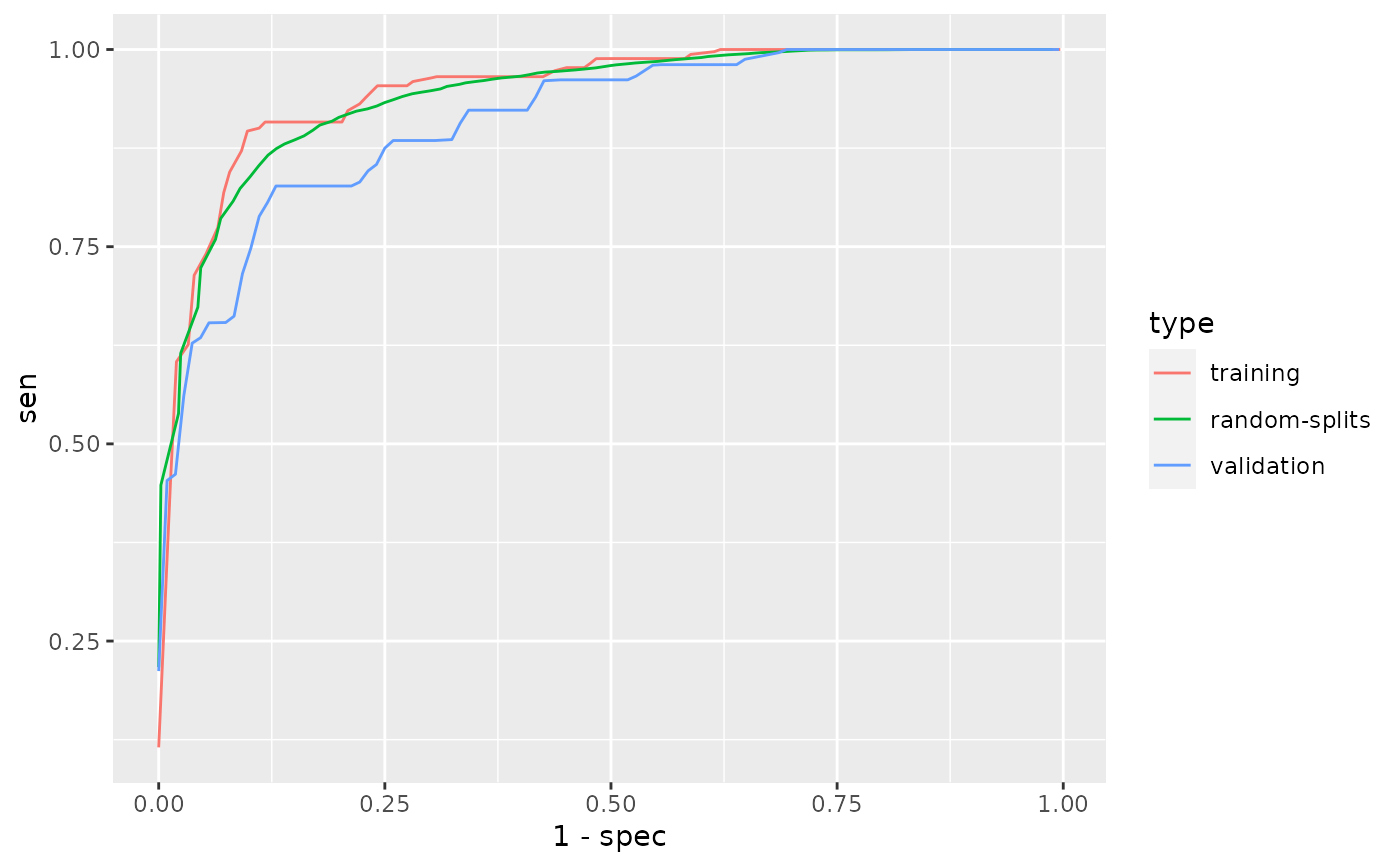
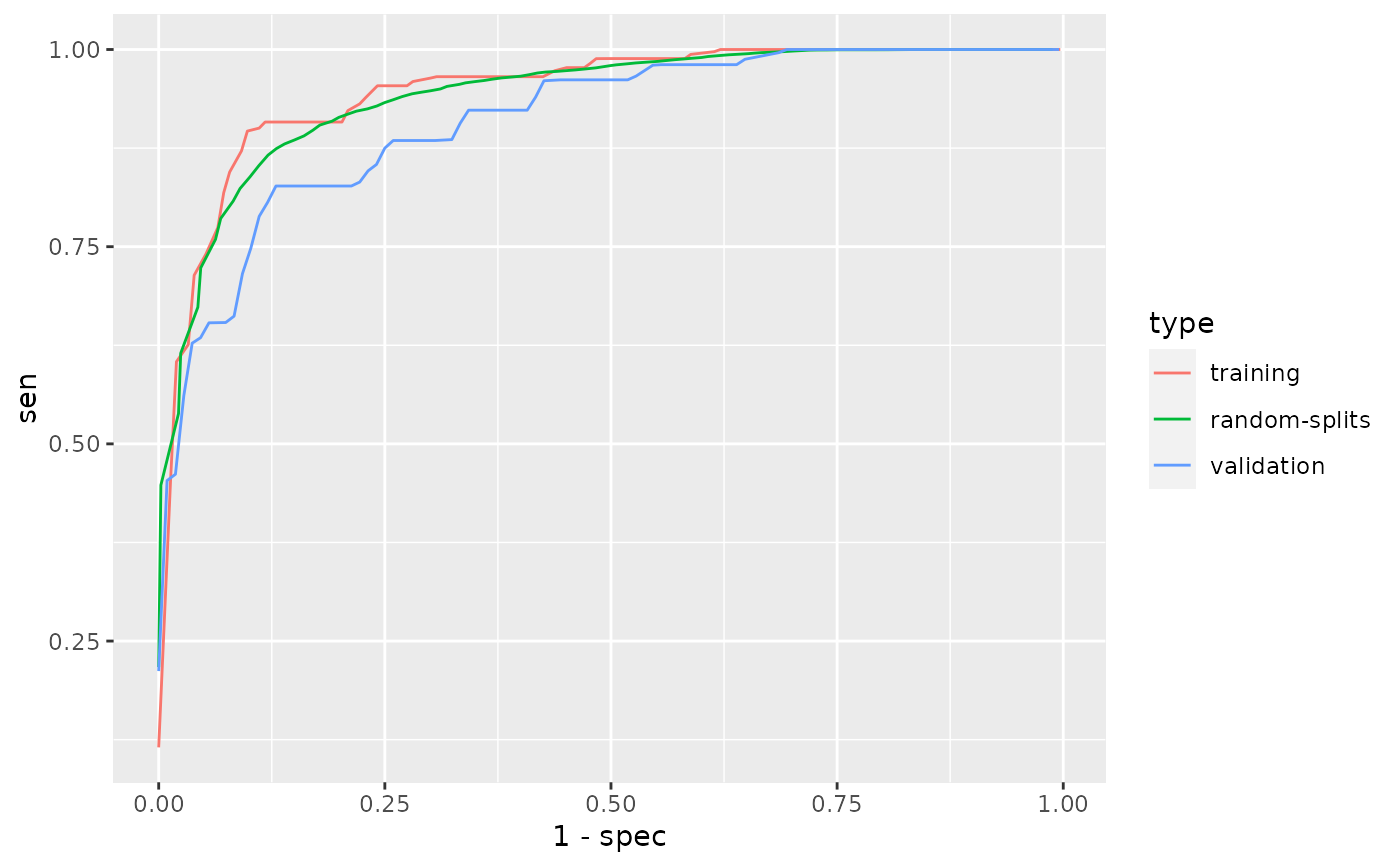
# Simulate an EHR dataset size <- 2000 latent <- rgamma(size, 0.3) latent2 <- rgamma(size, 0.3) ehr_data <- data.frame( ICD1 = rpois(size, 7 * (rgamma(size, 0.2) + latent) / 0.5), ICD2 = rpois(size, 6 * (rgamma(size, 0.8) + latent) / 1.1), ICD3 = rpois(size, 1 * rgamma(size, 0.5 + latent2) / 0.5), ICD4 = rpois(size, 2 * rgamma(size, 0.5) / 0.5), NLP1 = rpois(size, 8 * (rgamma(size, 0.2) + latent) / 0.6), NLP2 = rpois(size, 2 * (rgamma(size, 1.1) + latent) / 1.5), NLP3 = rpois(size, 5 * (rgamma(size, 0.1) + latent) / 0.5), NLP4 = rpois(size, 11 * rgamma(size, 1.9 + latent) / 1.9), NLP5 = rpois(size, 3 * rgamma(size, 0.5 + latent2) / 0.5), NLP6 = rpois(size, 2 * rgamma(size, 0.5) / 0.5), NLP7 = rpois(size, 1 * rgamma(size, 0.5) / 0.5), HU = rpois(size, 30 * rgamma(size, 0.1) / 0.1), label = NA) ii <- sample.int(size, 400) ehr_data[ii, "label"] <- with( ehr_data[ii, ], rbinom(400, 1, plogis( -5 + 1.5 * log1p(ICD1) + log1p(NLP1) + 0.8 * log1p(NLP3) - 0.5 * log1p(HU)))) # Define features and labels used for phenotyping. data <- PhecapData(ehr_data, "HU", "label", validation = 0.4) data#> PheCAP Data #> Feature: 2000 observations of 12 variables #> Label: 139 yes, 261 no, 1600 missing #> Size of training samples: 240 #> Size of validation samples: 160# Specify the surrogate used for # surrogate-assisted feature extraction (SAFE). # The typical way is to specify a main ICD code, a main NLP CUI, # as well as their combination. # The default lower_cutoff is 1, and the default upper_cutoff is 10. # In some cases one may want to define surrogate through lab test. # Feel free to change the cutoffs based on domain knowledge. surrogates <- list( PhecapSurrogate( variable_names = "ICD1", lower_cutoff = 1, upper_cutoff = 10), PhecapSurrogate( variable_names = "NLP1", lower_cutoff = 1, upper_cutoff = 10)) # Run surrogate-assisted feature extraction (SAFE) # and show result. feature_selected <- phecap_run_feature_extraction( data, surrogates, num_subsamples = 50, subsample_size = 200) feature_selected#> Feature(s) selected by surrogate-assisted feature extraction (SAFE) #> [1] "ICD1" "ICD2" "NLP1" "NLP3"# Train phenotyping model and show the fitted model, # with the AUC on the training set as well as random splits. model <- phecap_train_phenotyping_model( data, surrogates, feature_selected, num_splits = 100)#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' valuesmodel#> Phenotyping model: #> $lasso_bic #> (Intercept) ICD1 NLP1 HU ICD2 NLP3 #> -5.5078655 1.8891308 1.3883836 -0.5058757 0.0000000 0.0000000 #> #> AUC on training data: 0.948 #> Average AUC on random splits: 0.941# Validate phenotyping model using validation label, # and show the AUC and ROC. validation <- phecap_validate_phenotyping_model(data, model)#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' valuesvalidation#> AUC on validation data: 0.911 #> AUC on training data: 0.948 #> Average AUC on random splits: 0.941#> Warning: Use of `df$value_x` is discouraged. Use `value_x` instead.#> Warning: Use of `df$value_y` is discouraged. Use `value_y` instead.# Apply the model to all the patients to obtain predicted phenotype. phenotype <- phecap_predict_phenotype(data, model) # \donttest{ # A more complicated example # Load Data. data(ehr_data) data <- PhecapData(ehr_data, "healthcare_utilization", "label", 0.4)#> Error in PhecapData(ehr_data, "healthcare_utilization", "label", 0.4): Some 'hu_feature' are not found in the data: healthcare_utilizationdata#> PheCAP Data #> Feature: 2000 observations of 12 variables #> Label: 139 yes, 261 no, 1600 missing #> Size of training samples: 240 #> Size of validation samples: 160# Specify the surrogate used for # surrogate-assisted feature extraction (SAFE). # The typical way is to specify a main ICD code, a main NLP CUI, # as well as their combination. # In some cases one may want to define surrogate through lab test. # The default lower_cutoff is 1, and the default upper_cutoff is 10. # Feel free to change the cutoffs based on domain knowledge. surrogates <- list( PhecapSurrogate( variable_names = "main_ICD", lower_cutoff = 1, upper_cutoff = 10), PhecapSurrogate( variable_names = "main_NLP", lower_cutoff = 1, upper_cutoff = 10), PhecapSurrogate( variable_names = c("main_ICD", "main_NLP"), lower_cutoff = 1, upper_cutoff = 10)) # Run surrogate-assisted feature extraction (SAFE) # and show result. feature_selected <- phecap_run_feature_extraction(data, surrogates)#> Error in phecap_check_surrogates(surrogates, variable_list): Variable(s) main_ICD not foundfeature_selected#> Feature(s) selected by surrogate-assisted feature extraction (SAFE) #> [1] "ICD1" "ICD2" "NLP1" "NLP3"# Train phenotyping model and show the fitted model, # with the AUC on the training set as well as random splits model <- phecap_train_phenotyping_model(data, surrogates, feature_selected)#> Error in `[.data.frame`(frame, , surrogate$variable_names, drop = FALSE): undefined columns selectedmodel#> Phenotyping model: #> $lasso_bic #> (Intercept) ICD1 NLP1 HU ICD2 NLP3 #> -5.5078655 1.8891308 1.3883836 -0.5058757 0.0000000 0.0000000 #> #> AUC on training data: 0.948 #> Average AUC on random splits: 0.941# Validate phenotyping model using validation label, # and show the AUC and ROC validation <- phecap_validate_phenotyping_model(data, model)#> Warning: collapsing to unique 'x' values#> Warning: collapsing to unique 'x' valuesvalidation#> AUC on validation data: 0.911 #> AUC on training data: 0.948 #> Average AUC on random splits: 0.941#> Warning: Use of `df$value_x` is discouraged. Use `value_x` instead.#> Warning: Use of `df$value_y` is discouraged. Use `value_y` instead.# Apply the model to all the patients to obtain predicted phenotype. phenotype <- phecap_predict_phenotype(data, model) # }