Overview
Surrogate-guided ensemble Latent Dirichlet Allocation (sureLDA) is a label-free multidimensional phenotyping method. It first uses the PheNorm or MAP algorithm to initialize probabilities based on two surrogate features for each target disease, and then leverages these probabilities to guide the LDA topic model to generate phenotype-specific topics. Finally, it combines phenotype-feature counts with surrogates via clustering ensemble to yield final phenotype probabilities.
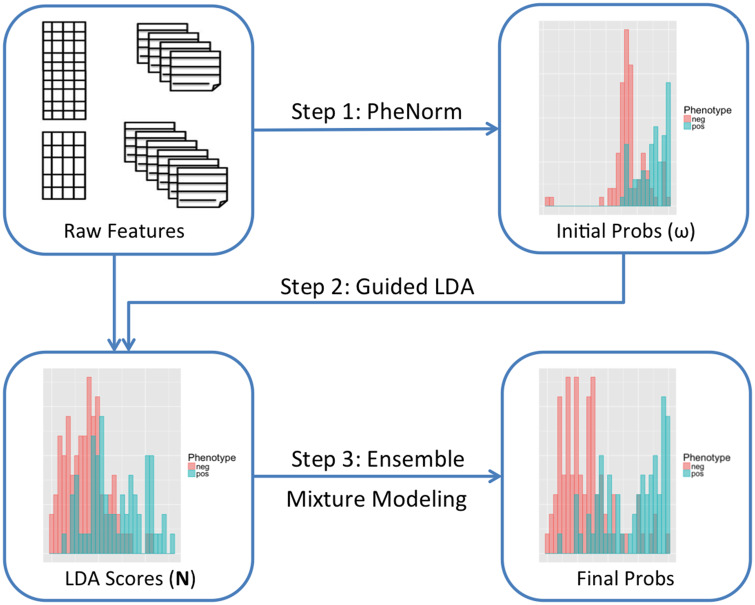
figure
Installation
Install stable version from CRAN:
install.packages("sureLDA")Install development version from GitHub:
# install.packages("remotes")
devtools::install_github("remotes/sureLDA")